Authors: Ahiwale S, Kapadnis B, Jagdale S
| Article Received: | 20/04/2021 |
| Received: | 28/05/2021 |
| Accepted: | 31/05/2021 |
Context: Multidrug-resistant strains of Klebsiella pneumoniae are ever emerging and creating a big challenge to healthcare worldwide. Therefore, there is a growing interest in potent lytic bacteriophages against multidrug-resistant Klebsiella pneumoniae.
Objective: This study reports isolation and characterization of a potent , water borne lytic phage of multidrug-resistant Klebsiella pneumoniae isolated from the hospital environment.
Methods: Pavana river water sample was used to isolate the phage against Klebsiella pneumoniae.
Results: Phage (vB_KpnM_KPP) isolated specific against Klebsiella pneumoniae from river water was identified as a member of the Myoviridae family, which is morphologically similar to the FC3 phage group. The adsorption rate constant was 4.7 ×10-10. Latent and eclipsed periods were 15 and 20 min, respectively, with the burst size of 120 phage particles per infected cell. The phage DNA size was 54 kb, and a proteome of 9 bands in the gradient gel was obtained. It is stable within a range of pH (5 to 10) and temperature (4 to 50 °C) range. As KPP, phage showed infectivity from pH 6 to 9 and temperature from 28 to 42 °C.
Conclusion: KPP is stable over a wide range of pH and temperature, indicating it s wide applications to control Klebsiella pneumoniae infections
Keywords: Klebsiella pneumoniae; Myovir idae; multidrug resistance; burstsize; stability; infectivity
Klebsiella pneumoniae is a Gram-negative, opportunistic bacterial pathogen belonging to the Enterobacteriaceae family. It has been associated with various types of infections, viz., hospital-acquired urinary tract infections, pneumonia, septicemia, soft tissue infections [1]. The infections caused by Klebsiella pneumoniae have been well documented n United States [2] and India [3]. Multidrug-resistant strains of Klebsiella pneumoniae are ever emerging. Recently, World Health Organization (WHO) has also warned regarding the emergence of multidrug-resistant bacteria worldwide and their big challenge to healthcare [4]. These multidrug-resistant bacteria are very hard to eradicate using available antibiotics. The extensive use of antibiotics has led to the development of multidrug-resistant strains of Klebsiella spp. About 80 % of nosocomial infections in immune-compromised patients are caused by multidrug-resistant strains of Klebsiella pneumoniae[5]. In one study, it has been found that over 60 % of strains of Klebsiella pneumoniae from the clinical cases were resistant to chloramphenicol and tetracycline [6]. Cephalosporin resistant strains of Klebsiella pneumoniae have been associated with increased morbidity and mortality in hospitalized patients [7]. Nosocomial infections caused by ESBL producing Klebsiella pneumoniae strains have been reported in Europe [8], United States, and South America [9]. The study in France showed plasmidmediated ESBL production in K. pneumoniae. The study carried out in the United States of America during 1998 - 2010 reported that the antimicrobial drug resistance in K. pneumoniae has increased for every antimicrobial class studied except tetracyclines and ciprofloxacin [11]. The biofilm formed by wild type Klebsiella pneumoniae resisted killing by ampicillin and ciprofloxacin was reported Anderl et al [12].
From the above reports, it is clear that Klebsiella pneumoniae has now become r esistant to almost all antibiotics available. therefore, there is a growing interest in lytic bacteriophages as an alternative for solving conventional antibiotic problems. Phages are highly specific against their host bacteria and are unable to kill the normal microflora; they are highly reproducible with no side effects [13, 14, 15]. Phages can be used as a potential therapeutic agent as they are host specific and lyse target bacteria efficiently [16]. Numerous lytic phages specific to multidrug-resistant Klebsiella pneumoniae have been isola ted and char acter ized. The majority of these are tailed phages, which belonged to families Myoviridae, Siphoviridae, and Podoviridae of or der Caudovirals [17]. This study reports a detailed characterization of indigenous, waterborne bacteriophage targeted against hospital isolate of Klebsiella pneumoniae.
Klebsiella pneumoniae was isola ted fr om the hospital environment on Mac Conkey’s (HiMedia) and Eosine Methylene Blue (HiMedia) agar media. All the suspected colonies were identified based on the identification recommended by Bergy’s Manual of Determinative Bacteriology18 that identifies bacteria based on morphological, cultural, and biochemical characteristics. Klebsiella pneumoniae was screened for its sensitivity to different antibiotics. Kirby-Bauer’s modified disc diffusion technique was used to check its antibiotic resistance pattern on the Muller-Hinton agar medium (HiMedia).
Lytic phages specific to multidrug-resistant Klebsiella pneumoniae wer e isolated fr om the Pavana river surface water, Pune, India. The water sample was collected in 250 ml sterile screw capped bottles. Further, the water sample was filtered through a sterile Nitrocellulose membrane (0.20 µm, Porafil @ NC from Machery-Nagel, Dueren, Germany). Then, phages were isolated from the collected filtrate by the double agar layer plaque method [19]. The mid-log phase culture (O.D = 0.57) of Klebsiella pneumoniae (0.5 ml) and the filtrate (0.5 ml) were mixed in 4 ml sterile soft agar (0.6 % w/v agar-agar, Qualigen Fine Chemicals Pvt. Ltd.) and then poured onto sterile nutrient agar medium. Plates were incubated at 37° C for 24 h and checked for the presence of plaques.
A single plaque (5 mm in diameter) was selected and transferred into sterile phage broth (250 ml) with 0.5 ml mid-log phase culture of Klebsiella pneumoniae. The fla sk was incubated at 37 °C for 24 h without agitation. Then, the contents were centrifuged, and the supernatant was filtered through a nitrocellulose membrane filter (pore size 0.20 µm). The lysate with enriched phage was stored at 4 °C. The phage titer in the lysate was determined by the double agar layer plaque method. Klebsiella pneumoniae phage hereafter was named KPP.
The plaque morphology of KPP was studied on nutrient agar and Mac Conkey’s agar media. An aliquot (1 ml) of phage lysate (2.4 × 108 pfu ml-1) was mixed with 1 ml of the mid-log phase culture, and then 0.1 ml of this mixture was spread onto the surface of the respective medium. The plates were incubated at 37 °C, observed at an interval of 3 h throughout the development of plaques. The oneway ANOVA test was used to evaluate the effect of nutrient media on the nature of plaques [20].
hage morphology was studied by sedimenting phage particles for 60 min using a Beckman J2-21 centrifuge (Beckman Instruments, Inc., Palo Alto, USA). Phages were washed twice with 0.1 M ammonium acetate buffer (pH 7.0), stained with 2 % phosphotungstate (pH 7.2), deposited on carboncoated Formvar films, and examined under a Philips EM 300 electron microscope at a magnification of 297,000 X.21

The cross infectivity study was carried out on different bacteria viz., Citrobacter koseri (MTCC 1657), Enterobacter aerogenes (MTCC 111), Escherichia coli (MTCC1678), Klebsiella pneumoniae (MTCC 39), Pseudomonas aeruginosa (MTCC 424), Proteus vulgaris (MTCC744), Salmonella Paratyphi A (MTCC 735), Salmonella typhimurium (MTCC 98), Shigella sonnei (MTCC 2957), and Vibrio cholerae (MTCC 3906). Cultures were obtained from the Institute of Microbial Type Culture Collection (IMTECH), Chandigarh, India. phase culture was mixed with 4 ml of soft agar (0.6 % w/v) and then poured onto a sterile nutrient agar plate. Once the overlay was solid and dry, a volume of 100 μl of phage lysate (2.4 x 108 pfu ml-1) was deposited at the center of each plate. Plates were incubated at 37 °C and examined for plaques after 6-10 h [22]. A clear zone in the bacterial lawn was recorded as complete lysis.
A mid-log phase culture of Klebsiella pneumoniae (OD650 = 0.57) (9 ml) grown in phage broth separately was infected with 1 ml of KPP (2.5 × 1010 pfu ml-1) and introduced into sterile 100 ml flasks equilibrated at 37 °C at time zero (t=0). The flasks were incubated at 37 °C in a shaker water bath at 160 rpm for one h. At one min interval, 50 µl aliquot was withdrawn from each flask and transferred into the two separate tubes containing 950 μl phage broths, supplemented with 5-6 drops of chloroform under cold conditions for 10 min. The tube contents were mixed thoroughly on a cyclomixer, serially diluted in phage broth, and then plated on sterile nutrient agar plates. Plaques were counted after overnight incubation at 37 °C.
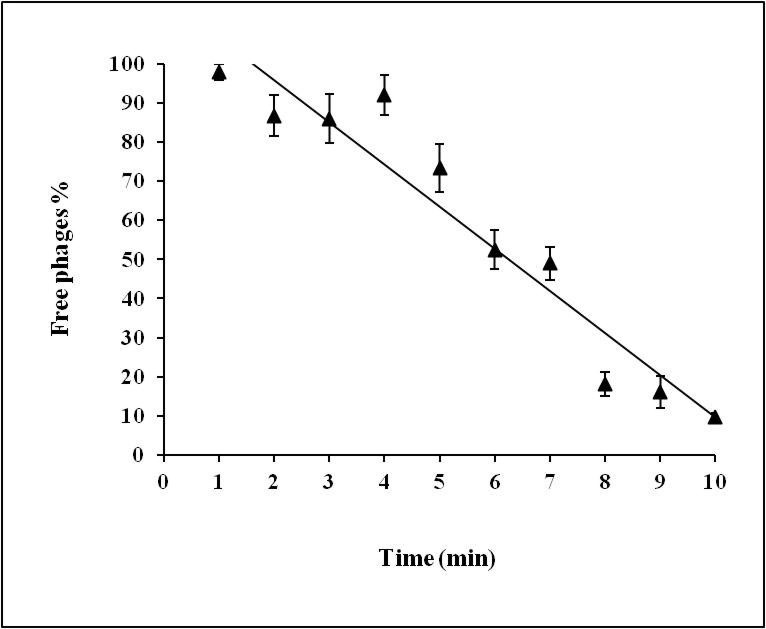
A one step growth curve was constructed as described by Hyman and Abedon23 with few modifications. riefly, 9 ml of the mid-log phase culture of Klebsiella pneumoniae (OD650 = 0.57) estimated on spectrophotometer UV 1800 (Shimadzu, Asia Pacific PTE Ltd., Singapore) was mixed with 1ml of KPP (2.5 × 1010 pfu ml-1) in a 100 ml flask (with MOI of 0.19). Phages were allowed to adsorb for 10 min at 37 °C. The mixture was then centrifuged (10,000 × g, 20 min, 4 °C), the pellet formed was resuspended in a 10 ml fresh phage broth medium. Two aliquots of the suspension (0.1ml each) were withdrawn at 5 min intervals throughout 1 h. One aliquot was transferred to a tube containing 0.9 ml of sterile phage broth, and the second aliquot was transferred into a tube containing 0.9 ml of phage broth with chloroform (1 % v/v) kept on ice. The plaque forming units (pfu ml-1) in each tube were determined.
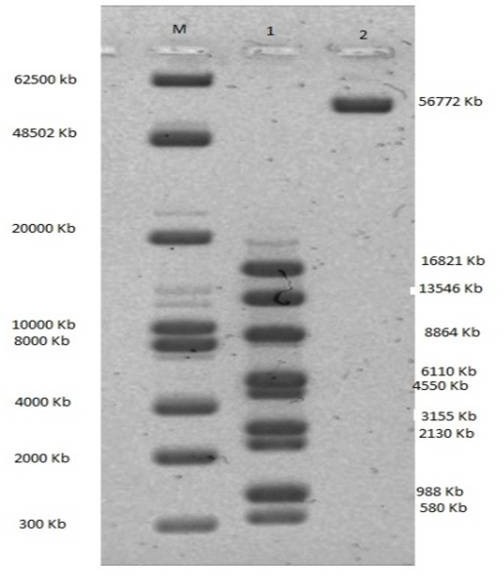
DNA from KPP phage was extracted as described by Ausubel et al.24 and purified by cesium chloride density gradient centrifugation as mentioned by Davis et al.25 using a Hitachi 55P Ultracentrifuge (Hitachi Koki Co. Ltd., Tokyo, Japan) at 64,000 × g for 24 h. Phage DNA was digested with EcoRI and Hind III restriction enzymes according to the manufacturer's instructions (Fermentas International Inc., Glen Burnie, MD, USA). The reaction mixture comprised Phage DNA 20 µl, 10X assay buffer 2.5 µl, nuclease free water 1.5 µl and 1 µl each of the restriction enzyme according to the manufacturer's instructions (Fermentas International Inc., Glen Burnie, MD, USA). The reaction mixture was incubated at 37 °C for 1 h in the water bath. Molecular weights of fragments were determined using an electrophoresis unit (BioEra Life Sciences Pvt. Ltd., Pune, India) at 100 V for 9 h on a 0.5 % agarose gel. A broad range DNA molecular weight marker (BioEra Life Sciences Pvt. Ltd., Pune, India) was used for control. Alpha Imager Software (BioEra Life Sciences Pvt. Ltd., Pune, India) was used to determine the molecular weight of the products.
The purified KPP phage was used for the extraction of proteins. The proteins were concentrated by precipitating using 10 % PEG 8000 after incubating at 4 °C overnight and then centrifuged at 12,000 × g for 25 min. The supernatant was discarded, and SM buffer (pH 7.5) was added to the glazy pellet. Further, phage proteins were concentrated with an Amicon kit (Millipore India Pvt. Ltd, Bangalore, India) using a 3 kDa cut-off membrane. 12 % SDSPAGE gel was prepared and loaded with phage protein (100 µg ml-1) with a standard molecular weight marker [26]. Protein bands were visualized after staining with Coomassie dye G-250 (Sigma-Aldrich, Bangalore, India).
The stability of KPP at varied pH 1-14 (0.05 mol l-1 citrate buffer: pH 4, 5 and 6; 0.05 mol l-1 phosphate buffer: pH 7, 8 and 9 and 0.05 mol l- of Tris-glycine buffer of pH 10, 11, and 12) was determined. 1 ml of the KPP (2.5 × 1010 pfu ml-1) lysate was suspended in 9 ml of buffers, respectively. All the tubes were incubated for 1 h at room temperature. Contents of the tubes were serially diluted in SM buffer (pH 7.5), and plaque forming units (pfu ml-1) in each tube were determined.
The stability of KPP at different temperatures (4, 10, 20, 30, 37, 40, and 50 °C) was determined. 1 ml of KPP lysate (2.5 × 1010 pfu ml-1) were incubated at the selected temperatures for 1 h, respectively. Contents of the tubes were serially diluted in SM buffer (pH 7.5), and plaque forming units (pfu ml-1) in each tube were determined.
The pH value from 4-10 was selected for the infectivity study of phage KPP. The mid-log phage culture (5 h) of Klebsiella pneumoniae (0.5 ml) was mixed with KPP lysate (2.5 × 1010 pfu ml-1) (0.1 ml) in a sterile tube (4 ml) containing soft agar (0.6 % w/v) and then plated onto sterile nutrient agar medium plates (varying in pH values from 4-10). Plates were incubated at 37 °C for 24 h and then were observed for plaques.
The temperature ranging from 0-42 °C was selected for the infectivity study of phage KPP. The mid-log phage culture (5 h) of Klebsiella pneumoniae (0.5 ml) was mixed with KPP lysate (2.5 × 1010 pfu ml1) (0.1 ml) in a sterile tube (4 ml) containing soft agar (0.6 % w/v) and then plated onto sterile nutrient agar medium plates. Plates were incubated at different temperatures (0-42 °C) for 24 h and then were observed for plaques. Bacteriophage was enriched for 24 hrs, the content was centrifuged at 6000 × g for 15 min, and the supernatant was collected. The supernatant was filtered through 0.22 micron syringe filters, and at least 3 mL of phage filtrate was collected for each bacterial host. The filtrate was stored at -20°C until further use.
Cu ; Cefuroxime, OF; Ofloxacin, T; Tetracyclin, G; Gentamycin, Ac; Amoxyclave, DO; Doxycycline-HCl, Cl; Ceftrioxone, C;Chloramphenicol, A; Ampicillin, CO; CO-trimoxazole, OF; Ofloxacin; ‘R’: Resistant, ‘I’: Intermediate
The single phage was isolated against Klebsiella pneumoniae and was named vB_KpnM_KPP. KPP plaque was clear but was surrounded by a turbid hallo. The average plaque diameter of KPP was 5 mm, and the average number of phage particles per plaque was 5 × 105 pfu/plaque on nutrient agar and Mac Conkey’s agar media. There was no significant difference in the average plaque diameter value and the average number of phage particles per plaque on nutrient agar and Mac Conkey’s agar media.
Transmission electron microscopy (TEM) revealed that KPP belongs to the Myoviridae family that resembles Citrobacter FC3 phage. The phage has a head of 85.5 nm with an extended tail of 95×17 nm and a contracted sheath of 50×20 nm. The phage has no neck, base plate, but the tail fibers are folded along the tail (Fig. 1).
Fig. 1: Transmission electron micrograph of phage KPP isolated against Klebsiella pneumoniae
KPP produced complete lysis (clear zone) on E. coli (MTCC 1678) and Klebsiella pneumoniae (MTCC 39). While it showed incomplete lysis (turbid zone) on Citrobacter koseri (MTCC 1657) and Shigella sonnei (MTCC 2957) and no lysis was obser ved on the other bacteria used for the cross infectivity study (Table 2). The hospital isolate of Klebsiella pneumoniae was isolated and checked for its sensitivity towards different antibiotics. The isolate showed resistance to almost all antibiotics.
KPP phage showed fast adsorption to its host. The number of free phages was 50 % in the first 6 min and declined to 9 % after 10 min. The affinity of KPP phage for Klebsiella pneumoniae, i.e., the adsorption rate constant was 4.7×10 -10 phage particles cell -1 min -1 (Fig. 2). The latent period of KPP was 15 min, and the burst size was 120 pfu infected cell -1 (Fig.3).
The genomic DNA of the KPP phage was digested with Eco RI and Hind III. Nine bands and one band, respectively, were visible and indicated an approximate size of 56.8 kb.
Restriction digestion pattern also revealed only one band after digestion with Hind III indicating no restriction site for Hind III on the KPP phage genome (Fig.4).
Total nine bands were revealed in SDS-PAGE in protein profiling of phage KPP. A protein band with molecular size 143.2 kDa was the highest, while 10.3 kDa was the lowest protein (Fig.5).
Fig. 5 KPP phage protein profile. Lane 1: High molecular weight protein marker (range 3-205 kDa). Lane 2: P r ot ein p r ofile of KPP p hage
KPP was stable within a wide pH range, 6 to 10. The stability was 100% at pH 7 to 9. However, the stability decreased to 90 % at pH 6 and 9. The stability of KPP was 100 % in the temperature range of 4 to 30 ° C. Stability was decreased beyond 30 °C. Effect of pH and temperature on infectivity of KPP phage.
The optimum range of pH for KPP infection to its host was from pH 6 to 8 with 100 % infectivity. KPP phage showed 10 % infectivity at pH 4 while 65 % infectivity at pH 5. For pH 9, 90 % of infectivity was shown by the KPP phage. The optimum temperature for KPP infection to its host was 37 °C, whereas infectivity decreased to 95 % at 42 °C. Infectivity was 80 % and 40 % at 28 and 8 °C, respectively.
Characterization of bacteriophages is a mandatory step to explore them in various fields [27]. It has been observed that, amongst the phages studied so far, 96% of phages are tailed phages, and the remaining 3.7% are polyhedral, filamentous, and pleomorphic. the present study, phage isolated is a tailed phage. Bacteriophages specific for Klebsiella pneumoniae are widely spread in nature and can be isolated from freshwater environments. We observed that the plaques of phage KPP were surrounded by halos, indicative of bacterial cell decapsulation. This observation suggested the phage produced a depolymerase enzyme that could diffuse through the agar layer. Phage depolymerases, often a part of the tail spike or tail fiber, can degrade bacterial capsular polysaccharides into their component oligosaccharide units during infection. Capsular polysaccharide depolymerases have multiple applications, including therapeutic agents against bacterial pathogens28 and preventing or eradicating biofilms.29 Such morphological feature of plaque is observed in the case of the FC3 phage group. The plaque features of KPP are similar to the FC3-9 phage of Citrobacter intermedius C3.30 The halo's presence might suggest the production of the soluble phage enzymes, e.g., polysaccharide depolymerases, as indicated by Huges et al.31 It has also been reported that FC3-9 phage shows capsular depolymerase activity.30 Polysaccharide depolymerase could be used for efficient capsular typing. The lytic phage and depolymerase have the potential as alternative therapeutic agents to antibiotics for treating K. pneumoniae infections, especially against antibiotic-resistant strains. Electron microscopic studies revealed that KPP has an icosahedral head of about 85.5 nm in diameter and a contractile tail of about 95 nm long. It belonged to the Myoviridae family. Morphologically, KPP resembles the FC3-9 phage of Citrobacter intermedius with a capsid of about 80 nm and a tail about 110 nm long.30 FC3 phages are reported in Klebsiella pneumoniae and Citrobacter spp.30,32 Bacteriophage FC3-9 is one of the several FC3 group phages of Klebsiella pneumoniae C3 that is reported. Mutants resistant to these bacteriophages are also isolated and found to be devoid of lipopolysaccharides O antigens.33 FC3 phage members require capsular receptors (lipopolysaccharides) for their binding.34 Therefore, it can be concluded that the host (Klebsiella pneu
Klebsiella pneumoniae showed 98.5-99.7% of adsorption in 5 min while KP 15 was able to adsorb only 75 %. Similarly, a myovirus (vBKpneM-Isf48) adsorbed up to 78 % in 4 min, and a siphovirus (vB_KpnS_Teh.1) adsorbed around 99 %, which were isolated against K. pneumoniae. The one step gr owth cur ve of KPP showed that the latent period is 15 min and the burst size is 120 pfu infected cell -1. These results contradict the previous works of FC3 phages where the range of latent period for FC3-1 to FC39 was 30-50 min, and the burst size was similar to FC3-4 phage. Further, Kęsik-Szeloch et al. reported that the latent period of siphovirus and podovirus was 15 min with a burst size of ~50-60 pfu infected cell -1 while myovirus showed 25 min latent period with burst size 10-15 pfu infected cell 1. The podovirus KP8 of K. pneumoniae showed a latent period of 15 min and ~ 40 pfu infected cell burst size [39]. The siphovirus (vB_KpnS_Teh.1) showed a 40 min latent period and 35-40 pfu infected cell -1 burst size [38]. Peng et al [40]. isolated phage vB_KleS-HSE3 (Siphovirius) against multidrug-resistant Klebsiella pneumonia, showed a latent period of 30 min with 277 pfu infected cell burst size. The restriction digestion pattern indicated that KPP DNA is resistant to Eco RI's digestion and sensitive to Hind III restriction enzyme. The genome size of KPP is 57 kb that is found to e close to the FC3-11 phage of K. pneumoniae genome size of 48-50 kb [35]. This newly isolated K. pneumoniae phage (KPP) shows pr oper ties similar to the FC3-9, FC3-11, FC3-4 phages that belong to the Myoviridae family and the φKMV-like virus particle of the Podoviridae family, indicating the genetic evolution. The proteome analysis revealed a total of 9 bands on the gel, with thee prominent protein bands viz., 143.2, 90.8, and 60 kDa and four less prominent bands viz., 42, 32, 18 kDa were detected. Similar proteins were also detected (16, 30, 33 90 kDa) in FC3-11phage of K. pneumoniae [35]. Similar proteins (94, 62, 42, 30, 20 kDa) were also found in the φKMV-like virus particle of K. pneumoniae that belongs to the Podoviridae family [41]. Morozova et al [39]. reported that 9 protein bands were revealed using SDS-PAGE analysis. This indicates that these proteins may be the common structural proteins in these phages. Storage stability is an essential parameter to apply phages as a biocontrol agent in the environment. KPP is found to be stable over a wide range of pH (6 to 10) and temperature (4 to 40 °C), indicating their wide applications to control K. pneumoniae infections. Similarly, in support of this, phages showed wide stability from pH 5-8 and temperature 4 to 50 °C [37], phage vB-KpneM-Isf48 showed pH stability from 6-9, and temperature 30 and 40 °C, phage vB_KpnS_Teh.1 showed pH stability at 7 and temperature at 37 °C, and phage vB_KleS-HSE3 showed pH stability from 5-11 and temperature 4 to 50 °C [40]. Besides, phage KPP showed 100 % infectivity at pH (6-8) and temperature (37 C), respectively.
The newly isolated phage of Klebsiella pneumoniae has many unique features such as a short generation ime and high stability over a wide range of pH and temperature, making it a promising biocontrol agent for drug-resistant strains of Klebsiella pneumoniae.
Authors have declared no conflict of interest.
The authors acknowledge the late Dr.H.W.Ackermann, Canada, for the TEM image of the KPP phage.
Nil
1. Podschun R, Ullmann U. Klebsiella spp. As nosocomoal pathogens: Epidemiology, taxonomy, typing methods and pathogenicity factors. Clin Microbiol Rev. 1998; 11:589- 603.
2. Graybill JR, Marshall IW, Chareche P,Wallace CK, Melwin VK. Nosocomial pneumonia: A continuing major problem. Am Rev Respir Dis. 1973; 108:11301140.
3. Mathur NB, Khalib A, Sarkar R, Puri RK. Mortality in neonatal septicemia with involvement of mother in management. Ind J Pediatri. 1991; 28:1259-1264.
4. Young-soo S. World Health Organization, Western pacific region, Press release. 2011.
5. Chhibber S, Kaur S, Kumari S. Therapeutic potential of bacteriophage in treating Klebsiella pneumoniae B5055mediated lobar pneumonia in mice. J Med Micribiol. 2008; 57:1508-1513.
6. Sikarwar AS, Batra HV. Prevalence of antimicrobial drug resistant to Klebsiella pneumoniae in India. Int J Biosci Biochem Bioinform. 2011; 1:211-215.
7. Dean AD, Dean AJ, Burton AH, Dicer RC. EpiInfoversion 5: a word processing, databasem and statistics systems for epidemiology on microcomputers. In Epi Info-version 5: a word processing, databasem and statistics systems for epidemiology on microcomputers. 1990, pp. 384-384.
8. Arlet G, Rouveau M, Casin I, Bouvet PJM, Lagrange PH, Phillippon A. Molecular epidemiology of Klebsiella pneumoniae strains that produce SHV4- β Lactamase and which were isolated in 14 French hospitals. J Clin Microbiol. 1994; 32:2553-2558.
9. Jarher MH, Fournier NG, Phillipon A. Extended broad spectrum β-lactamases conferring trasnferable resistance to newer beta lactamase agents in Enterobacteriaceae hospital prevalence and susceptibility pattern. Clin Infect Dis. 1998; 10: 867-878.
10. acoby GA, Medeiros AA. More extended-spectrum betalactamases. Antimicrob Agents Chemother. 1991; 35:1697.
11. Sanchez GV, Master RN, Clark RB, Fyyaz M, Duvvuri P, Gupta E, Bordon J. Klebsiella pneumoniae antimicrobial drug resistance, United states 1998-2010. Emerg Infect Dis. 2013; 19:133-135.
12. Anderl JN, Franklin MJ, Stewart PS. Role of antibiotic penetration in K. pneumoniae biofilm resistance to ampicillin and Ciprofloxacin. Antimicrob Agents Chemother. 2000; 44:1818-1824.
13. O'Flaherty S, Ross RP, Coffey A. Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol Rev. 2009;33:801-819.
14. Carson L, Gorman SP, Gilmore BF. The use of lytic bacteriophages in the prevention and eradication of biofilms of Proteus mirabilis a nd Escherichia coli. FEMS Immunol Med Microbiol. 2010; 59:447-455.
15. Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM. Phage treatment of human infections. Bacteriophage. 2011;1:66-85.
16. Morozova VV, Vlassov VV, Tikunova NV. Applications of bacteriophages in the treatment of localized infections in humans. Front Microbiol. 2018; 9:1696.
17. Sasani MS, Eftekhar F, Hosseini M. Isolation and Characterization of a Klebsiella pneumoniae Specific Lytic Bacteriophage from a Hospital Waste-water Treatment Plant. J Med Microbiol Infect Dis. 2019; 7:6-11.
18. Bergey DH, Holt JG, Krieg P (1994) Bergey's manual of determinative bacteriology. Williams and Wilkins, Baltimore, MD, USA.
19. Adams MH. Bacteriophages. Interscience Publishers Inc, New York, 1959.
20. Khan IA, Khanum A. Fundamentals of biostatistics. Ukaaz Publications, Hyderabad, India, 2008.
21. ckermann HW. 5500 Phages examined in the electron microscope. Arch Virol. 2007; 152:227-243
22. Manchester LN. Characterization of a bacteriophage for Carnobacterium divergens NCFB 2763 by host specificity and electron microscopy. Lett Appl Microbiol. 1997; 25: 401-404.
23. Hyman P, Abedon ST. Practical methods for determining phage growth parameters. In: Bacteriophages, Humana Press, 2009, pp 175-202.
24. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. John Wiley & Sons, New York, 1999.
25. Davis RW, Botstein D, Roth JR. Advanced Bacterial Genetics: A Manual for Genetic Engineering. CSH Laboratory, Cold Spring Harbor, NY, 1980.
26. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970; 227: 680–685.
27. Ackerman HW. Phage classification and characterization. In: Clokie MRJ, Kropinski AM (Eds) Bacteriophages: methods and protocols, Volume I: Isolation, characterization and interactions. Humana Press, New York, 2009, pp 127-140.
28. Mushtaq N, Redpath MB, Luzio JP, Taylor PW. Prevention and cure of systemic Escherichia coli K1 infection by modification of the bacterial phenotype. Antimicrob Agents Chemother. 2004; 48: 1503–1508.
29. Verma V, Harjai K, Chhibber S. Structural changes induced by a lytic bacteriophage makes ciprofloxacin effective against older biofilm of Klebsiella pneumoniae. Biofouling. 2010; 26:729-737.
30. Camprubi S, Merino S, Benedi VJ, Tomas JM. Isolation and characterization of bacteriophage FC3-10 from Klebsiella spp. FEMS Micr obiol Lett. 1991 ; 67: 291-297.
31. Regue M, Tomas J, Pares R, Jofre J. Isolation and partial characterization of phages infecting Citrobacter intermedius. Ca n J Micr obiol. 1981; 5: 153-156.
32. Huges KA, Sutherland IW, Clarck J, Jones MV. Bacteriophage associated polysaccharide polymerases-novel tools for study of bacterial biofilms. J Appl Microbiol. 1998;85:583-590.
33. Enedi VJ, Ciurana B, Tomas JM. Isolation and characterization of Klebsiella pneumoniae uncapsulated mutants. J Clin Microbiol. 1989; 27:82-87.
34. Tomas JM, Jofre JT. Lipopolysaccharide-specific bacteriophage for Klebsiella pneumoniae C3. J Bacteriol. 1985;162:1276-1279.
35. Hernandez S, Alberti S, Rubires X, Merino S, Tomas JM, Benedi VJ. Isolation of FC3-11, a bacteriophage specific for the Klebsiella pneumoniae porin OmpK36, and its use for the isolation of porin- deficient mutants. Can J Microbiol. 1995;41:399-406.
36. Bielke L, Higgins S, Donoghue A, Donoghue D, Hargis BM. Salmonella host range of bacteriophages that infect multiple genera. Poult Sci. 2007; 86:2536-2540.
37. Kęsik-Szeloch A, Drulis-Kawa Z, Weber-Dąbrowska B, Kassner J, Majkowska-Skrobek G, Augustyniak D, Łusiak-Szelachowska M, Żaczek M, Górski A, Kropinski AM. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Vir ol J. 2013; 10:100.
38. Komijani M, Bouzari M, Rahimi F. Detection and characterization of a novel lytic bacteriophage (vB-KpneMIsf48) against Klebsiella pneumoniae isola tes fr om infected wounds carrying antibiotic-resistance genes (TEM, SHV, and CTX-M). Iran Red Crescent Med J. 2017; 19:e34475.
39. Morozova V, Babkin I, Kozlova Y, Baykov I, Bokovaya O, Tikunov A, Ushakova T, Bardasheva A, Ryabchikova E, Zelentsova E, Tikunova N. Isolation and characterization of a novel Klebsiella pneumoniae N4-like bacteriophage KP8. Viruses. 2019; 11:1115.
40. Peng Q, Fang M, Liu X, Zhang C, Liu Y, Yuan Y. Isolation and Characterization of a Novel Phage for Controlling Multidrug-Resistant Klebsiella pneumoniae. Microorganisms. 2020;8:542.
41. awa DZ, Mackiewicz P, Szeloch AK, Dziubinska EM, Dąbrowska BW, Jach AD, Augustyniak D, Skrobek GM, Bocer T, Empel J, Kropinski AM. Isolation and characterisation of KP34 - a novel 8KMV-like bacteriophage for Klebsiella pneumoniae. Appl Micr obiol Biotechnol. 2011;90:1333-1345.
Ahiwale S, Kapadnis B, Jagdale S . Characterization of lytic bacteriophage isolated against multidrug-resistant Klebsiella pneumoniae Int J Bacteriophage Res 2021:1:42-50