Authors: Prasanth M, Kandasamy E, Loganathan A, Madurantakam R.M, Sebastian L, Nachimuthu R.
| Article Received: | 24/12/2020 |
| Received: | 28/12/2020 |
| Accepted: | 02/01/2021 |
Context: Bacteriophages are one of the most promising alternatives to antibiotics in the treatment of bacterial infections.
Objectives: In this study, virulent (lytic) phages were isolated from different sample sources and morphologi-cally identified using Transmission Electron Microscopy.
Methods:From sewage water samples, more than 50 bacteriophages were isolated against Escherichia coli, Klebsiella, Enterobacter, Pseudomonas, Serratia and Staphylococcus. Escherichia phages were isolated from river water of the Ganges. The Citrophages were isolated from marine water samples and mycobacteriophages from soil samples within the vicinity of a hospital in India.
Results: The bacteriophages were isolated using the phage enrichment method, and virulent phages were identified using plaque morphology. For the analysis by electron microscopy, phage samples were prepared using three different methods; filtration, precipitation and purification. Here, the simple phage filtration method was found to give the best TEM results. For the phages we analysed, we found that staining with 1% [w/v] uranyl ace- tate was better than 2% phosphotungstic acid. The bacteriophages which may have therapeutic potential, were iso- lated from eight different bacterial genera, and belong to the Myoviridae, Siphoviridae and Podoviridae.
Conclusion: The sewage water samples are excellent sources for bacteriophages, and virulent phages can be rap- idly characterized using plaque morphology and TEM.
Keywords: Bacteriophages, Virulent phage, Lytic cycle, Plaque morphology, TEM analysis, Phage therapy
Bacteriophages are the most abundant organisms on Earth and an estimated 10 [31] phage particles on this planet. Phages are known to present in every environment in which bacteria exist, and there is at least one type of phage, more than one in most cases, to infect every strain of bacteria. With their ubiquity and diversity, phages play a profound role in determining bacterial diversity and evolution. One hundred years ago, Frederick Twort discovered bacteriophages, followed by the studies on its various applications. Phage therapy is the use of bacteriophages to treat bacterial infections in humans. Before the discovery of antibiotics, bacteriophages were the choice of treatment against bacterial infections such as diarrhoea.[3] But soon after the introduction of antibiotics, the use of bacteriophages in therapy was almost abandoned.[4] The overuse and mishandling of antibiotics led to the development of antibiotic-resistant bacteria which is one of the most worrisome healthcare problems. In the post-antibiotic era, it becomes necessary to combat antibiotic-resistant bacterial infections using alternative therapies as antimicrobial compounds are ineffective. Phage therapy receives renewed interest among phage researchers, and fundamental and applied studies on bacteriophages increased dramatically, recently also including clinical trials.[5]
The research on bacteriophages for clinical or biotechnological purposes gained increasing attention after the 2000s, especially the isolation of virulent bacteriophages for the treatment of bacterial infections, and the preparation of phage banks for personalized phage delivery.[6] Though bacteriophages and their proteins have a multitude of applications in different fields, for clinical use therapeutic phages are required to be characterized in much detail. Virulent phages undergo a so-called lytic cycle that differs from that of the temperate phages which are characterized by a lysogenic cycle. During this cycle, the phage genome integrates into the host genome, which can present as an advantage to the host as some prophages (i.e. genome integrated lysogenic phages) influence host behavior or virulence by encoding virulence factors such as toxins or antimicrobial resistance genes.[7,8,9] Phages suitable for therapeutic purposes are generally lytic, where the produced phage progeny is released by lysis (the destruction of the bacterial cell envelope) after their replication inside the host bacterium. With this specific quality, virulent phages can be used to kill the pathogenic bacteria inside the human system and more often phages are bacteria-specific. For therapy, phages that are strictly lytic are preferred to reduce the complication of horizontal gene transfer as lysogenic phages can transfer genes by transduction.[10] The majority of the therapeutic phages fall within the Order Caudovirales (tailed phages), and they are grouped into three morphological fami- lies, i) Myoviridae (long contractile tail with tail fibers), Siphoviridae (long non-contractile tail) and Podoviridae (short non-contractile tail). The recent International Committee on Taxonomy of Viruses (ICTV) classification grouped Caudovirales into nine families.[11]
Previous studies explain the importance of bacteriophage characterization for phage therapy.[12-16] With the increasing use of antibiotics during the coronavirus pandemic to treat or prevent secondary bacterial infections, we are facing another global threat, this time of widespread antibiotic resistance of pathogenic bacteria.[17] Phage therapy might be a possible solution to overcome such a crisis, and therefore has to be studied in detail. Our work outlines a simple and effective method for the identification of lytic bacteriophages with therapeutic potential (Fig.1).
The challenges prior to the deployment of phages for clinical therapy include the isolation and identification of virulent phages, their purification and identifying suitable storage and delivery conditions.[18-19] The microscopic analysis of phage morphology is an important characterization technique to identify the tailed phages which is often followed by genomic analysis. This study aimed to describe simple methods for the isolation and identification of virulent bacteriophages from the envi- ronmental sources, using plaque morphology and Transmission Electron Microscopic (TEM) analysis.
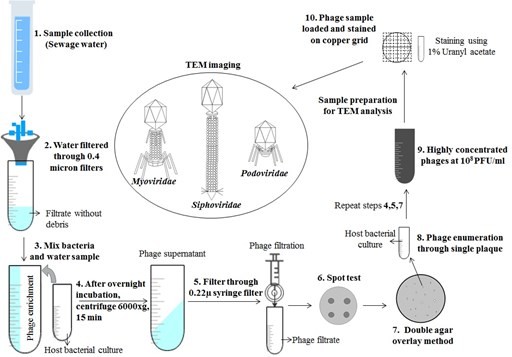
Figure 1: Schematic of virulent or lytic phage isolation and morphological characterization using transmission electron microscopic analysis.
For the isolation of bacteriophages, the samples were collected from Ganges river water, city of Varanasi, municipal sewage water treatment plants in Vellore, Chennai and Karur, iii) hospital sewage-waste water in Vellore and Chennai, soil samples within the hospital area at Government Vellore Medical College & Hospital, and marine water from the coastal regions of Ramanathapuram, Tamil Nadu. The collected water or soil samples were processed immediately to isolate bacteriophages or filtered using 0.45 micron filters and stored at 4°C.
The bacteria used for the isolation of bacteriophages include the clinical isolates of Escherichia coli, Klebsiella pneumoniae, Enterobacter cloacae, Pseudomonas aeruginosa, Serratia marcescens, Staphylococcus aureus, whereas, Citrobacter sp. was isolated from marine water samples, and Mycobacterium smegmatis mc2155. The bacteriophage isolation was performed using the phage enrichment method as follows; to a 1 mL of bacterial culture (OD600 =0.6) 9 mL of water samples were added and kept in an incubator shaker (120 rpm, 37°C) for 24 hrs.12 [Note: For soil samples, the phage enrichment method). Once the bacteria bacteriophage was enriched for 24 hrs, the content was centrifuged at 6000 × g for 15 min, and the supernatant was collected. The supernatant was filtered through 0.22 micron syringe filters, and at least 3 mL of phage filtrate was collected for each bacterial host. The filtrate was stored at -20°C until further use.
The phage filtrate was tested for bacteriophages using the spot test followed by the double agar overlay method. In the spot test method, lawn culture was prepared using the exponentially grown host bacterial culture on the Luria Bertani (LB) agar plate and 0.2-0.5 µL of respective phage filtrate was spotted on the bacterial lawn. The plates were allowed to dry for 15 min and incubated at 37°C for 15 hrs. The presence of clear spots indicates the antibacterial activity of phages. To further confirm the presence of phages, a double agar overlay method was performed. To a 200 µL of bacterial culture, 100 µL of phage filtrate was added, and the mixture was kept undisturbed for 10 min and 3 mL of molten soft agar (0.75% agar) was added, mixed well and poured on to the pre-prepared LB agar plates. The plates were dried and incubated at 37°C for 24 hrs. The appearance of plaques indicates the presence of bacteriophage and plaque size was measured after 24 hrs incubation.
For identification of mycobacteriophages, M. smegmatis mc2155 bacterial lawn culture (host bacterium) was prepared in the Middlebrook 7H10 agar plates supplemented with carbenicillin (50 µg/mL), cycloheximide (10 µg/mL) and 1mM CaCl2. Then, 10 μL of enriched phage filtrate was spotted and the plates were incubated at 37°C for 16-20 hrs. The appearance of clear spots indicated the presence of mycobacteriophages. For dou- ble agar overlay method, in a 15 mL centrifuge tube, 0.5mL M. smegmatis mc2155 cells and 50 µL of enriched phage lysate were added and incubated at room temperature for 20 min. To this, 4.5 mL of M7H10 soft agar was added and layered over a pre-prepared M7H10 agar plate. The plates were incubated at 37°C for 48 hrs and observed for plaque formation and plaque size was measured after 48 hrs incubation.
Morphological characterization of bacteriophages was performed using TEM imaging following previously established protocols.[12-14] For TEM analysis, the phage samples were prepared using three different methods. I) Highly concentrated phages at 108 PFU/mL were pre- pared without following any purification processes, II) the phages (108 PFU/mL) were precipitated using 10% polyethylene glycol (PEG 8000) and 1% sodium chlo- ride (NaCl) and used for TEM analysis, III) the phages (108 PFU/mL) were purified using sucrose gradient centrifugation and the purified phages were used for TEM analysis. In all the cases, the phages were highly con- centrated to enable easy TEM imaging. For TEM analysis, 2 µL of phage filtrate/precipitate/lysate was placed on the copper grid and allowed to settle for 10 min. The grid was dried to free the sample, and 1% [w/v] uranyl acetate or 2% [w/v] phosphotungstic acid (PTA) solu- tion was added. The grid was kept undisturbed for 30 sec, and the staining solution was cleared. The grid was washed thrice with distilled water to remove excess stain and died for 30 min. The copper grid with a negatively stained phage sample was visualized using TEM [FEI-TECNAI G2-20 TWIN, Bionand, Spain] at the VIT-TEM facility.
The bacteriophages against their host bacteria were isolated from the different sample sources. The strategy of a simple bacteriophage isolation and identification method, which we have used, is provided in Fig. 1. For the isolation of bacteriophages against the clinically relevant bacteria, water samples collected from sewage treatment plants and hospital waste water collected from the Indian cities of Chennai, Vellore and Karur were used. We were able to isolate more than 50 bacteriophages against E. coli, K. pneumoniae, E. cloacae, P. aeruginosa and S. marcescens from sewage water samples (Table 1). Marine water samples were used to isolate bacteriophages against the environmental bacteria, for example, Citrobacter causing aquaculture infections. For the isolation of mycobacteriophage, soil samples close to Government Vellore Medical College & Hospital were processed. Mycobacteriophages were mainly studied to tackle tuberculosis causing Mycobacterium sp.
The Pseudomonas phage PNR01 was isolated against P. aeruginosa and found to infect both virulent and non-virulent P. aeruginosa strains. A total of six phages were isolated against Pseudomonas, and only one phage isolated from sewage water collected in Chennai was found to have broad-host range activity. The isolated phage was found to infect 34/45 P. aeruginosa strains. The isolated Siphoviridae phage was found to form clear plaques (Fig.2), and the TEM image shows an icosahedral head of 57 ± 3.0 nm and a long non-contractile tail of about 255 ± 5.0 nm in length (Fig.2). The family Siphoviridae is the most abundant tailed-phage.[20] Sewage water samples present a highly valuable source of diverse microorganisms. TEM that should preferentially not belong to the Siphoviridae as many phages of this family exhibit a lysogenic lifecycle, and would require further testing e.g. the absence of genes that allow genome insertion.[21]
| Bacterial host | Sample used for phage isolation | Number of phages isolated | Characterized phages (#family) |
|---|---|---|---|
| Pseudomonas aeruginosa | Sewage water | 6 | 1 (Siphoviridae) |
| Serratia marcescens | Sewage water | 1 | 1 (Siphoviridae) |
| Staphylococcus aureus | Sewage water | 10 | 2 (Siphoviridae) |
| Citrobacter sp. | Marine water | 2 | 2 (Siphoviridae and Pod- oviridae) |
| * Mycobacterium smegmatis mc2155 | Soil sample from hospital | 3 | 2 (Siphoviridae) |
| Escherichia coli | Ganges River water, Va- ranasi Sewage water |
10 25 |
1 (Siphoviridae) 1 (Siphoviridae) |
| Klebsiella pneumoniae | Sewage water | 8 | 1 (Podoviridae) |
| Enterobacter cloacae | Sewage water | 5 | 1 (Myoviridae) |
* Characterized phages- number of phages having broad-host-range activity; #family- morphological differentiation based on TEM imaging. Samples collected: i) Ganges river water, city of Varanasi, ii) municipal sewage water treatment plants in Vellore, Chennai and Karur, iii) soil samples within the hospital area at Government Vellore Medical College & Hospital, and iv) marine water from the coastal regions of Ramanathapuram, Tamil Nadu.
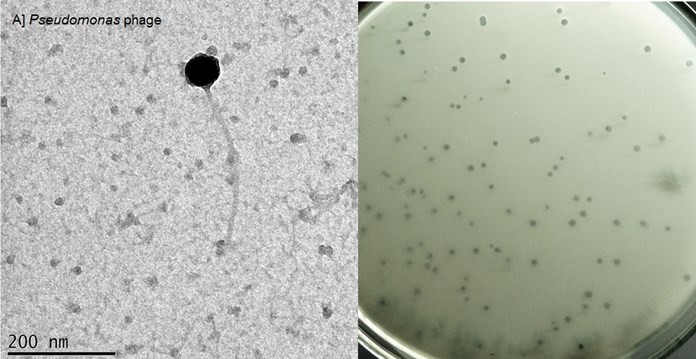
Figure 2: Transmission Electron micrographs of Pseudomonas phage (left) and the plaques formed on the double agar overlay plate (right).
The Serratia phage PNR007 was isolated from sewage samples collected in the Indian city of Vellore. Only one Serratia phage was isolated against pathogenic S. marcescens and found to infect all five S. marcescens strains tested. The isolated Siphoviridae phage is morphologically a tailed phage with an icosahedral head of 60 ± 5 nm and a long non-contractile, flexible tail of about 270 ± 5 nm (Fig.3). Only few studies on Serratia phages have been reported as the bacterium is not associated with severe disease outbreaks. However, infections by antibiotic- resistant S. marcescens are regularly reported.22,23 Up to date, less than ten Serratia phages have been reported, and the therapeutic characteristics of the Serratia phage PNR007 remain to be studied in detail.
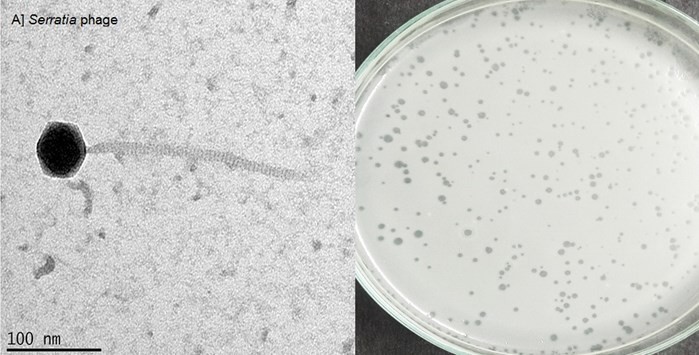
Figure 3: Transmission Electron micrographs of Serratia phage (left) and the plaques formed on the double agar overlay plate (right).
The Citrobacter phages, Citrophage MRM19 and Citrophage MRM57 were isolated from marine water samples collected near the coastal regions of Ramanathapuram, Tamil Nadu. The Citrophage MRM19 and Citrophage MRM57 were infecting Citrobacter werkmanii and Citrobacter amalonaticus, respectively.[14] The isolated bacteriophage Citrophage MRM19 (against C. werkmanii) showed a plaque morphology of small and irregular shapes (0.75 ± 0.2 mm), whereas Citrophage MRM57 (against C. amalonaticus) showed medium-sized, clear and circular plaques (2.0 ± 0.5 mm). Both the phages were found to have broad-host-range activity with Citrophage MRM19 infecting eight strains and
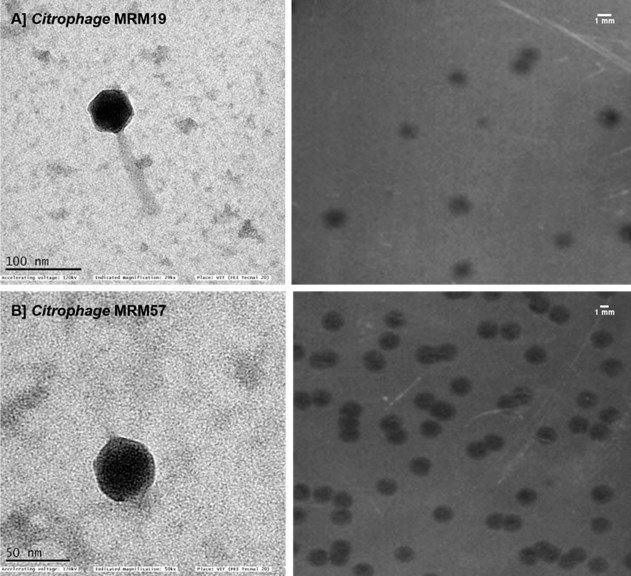
Figure 5: Transmission Electron micrographs of Staphylococcal phage (Top left and bottom left) and the plaques formed on the double agar overlay plate (Top right and bottom right).
Citrophage MRM57 infecting 15 strains. The morphological characterization revealed that both phages possess an icosahedral head with Citrophage MRM19 showing the typical features of a Siphoviridae phage with a head size of 60.7 ± 1.2 nm and a long non-contractile tail (120.0 ± 2.0 nm). Citrophage MRM57 belongs to the Podovir- idae with a head size of 48.0 ± 1.0 nm and a small tail size of 11.95 ± 1.92 nm (Fig.4). In particular for the aquaculture industry, infections by Citrobacter species are of great concern. The fish pathogen can cause massive economic loss in the commercial aquaculture industry across South Asia. Instead of using massive amounts of antibi- otics, the deployment of phages is promising.24 The in vivo efficacy of these phages in zebra fish (Danio rerio) infection model also showed prom- ising results to combat Citrobacter infections pub- lished previously.[14]
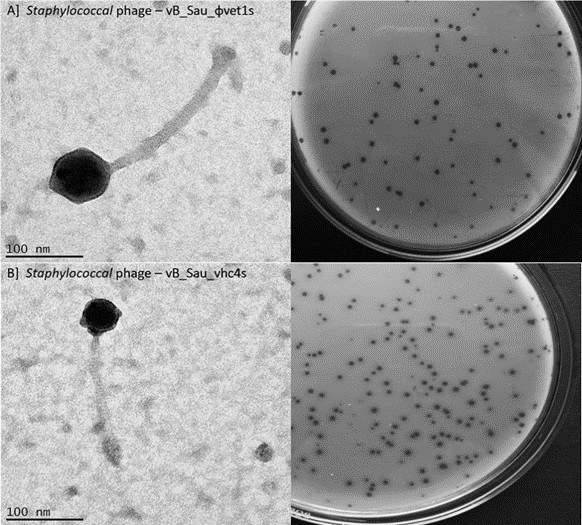
Figure 6: Transmission Electron micrographs of Mycobacterium Phage VIT001 and Mycobacterium phage VIT002 (Top left and bottom left) and the respective plaques formed on the double agar overlay plate (Top right and bottom right).
Staphylococcus aureus is an opportunistic pathogen that causes skin abscesses, endocarditis, osteomyelitis, pneumonia, and toxic shock syndrome.[25] The two staphylococcal phages (vB_Sau_ɸvet1s and vB_Sau_ɸvhc4s) were isolated against the clinical isolates of S. aureus from sewage water samples collected in Chennai and Vellore. Both phages exhibited a plaque size of approximately 4 ± 0.6 mm in double agar overlay, showing a clear plaque. TEM observation showed that both the phages belong to Si- phoviridae family. Phage vB_Sau_ɸ vet1s showed an icosahedral head of size 82 ± 0.5 nm and long non-contractile tail of about 214 ± 0.9 nm and another phage vB_Sau_ɸvhc4s showed an icosahedral head of size 47 ± 0.6 nm and long non-contractile tail of about 170 ± 2 nm (Fig.5). Both staphylococcal phages showed a broad host range of 83% (177/213) and 62% (132/213) respectively, similarly to S. aureus phage JD419.[26] Earlier studies using phages against S. aureus infections showed promising results against local and systemic infections.[27] The use of so-called phage cocktails -a mixture of different phages in a single solution- has the promising ability to cure complex bacterial infections also caused by biofilm-forming bacteria. In the light of MRSA infections becoming more common in the developing countries, the use of phages or phage cocktails containing staphylococcal phages becomes more likely.[28]
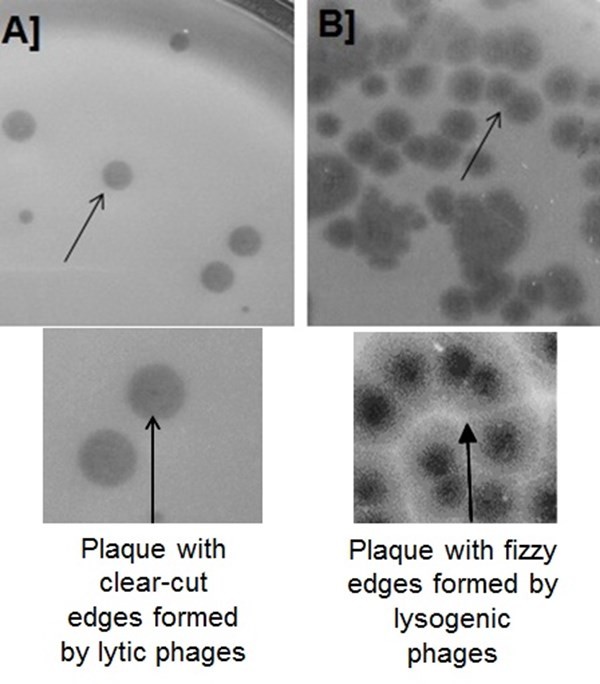
Figure 7: Plaque morphologies of the bacteriophages on the double agar overlay plate. A) A clear halo-plaques formed by the virulent phages and B) Turbid, bulls-eyed plaques formed by the temperate phages.
Tuberculosis (TB) is one of the deadliest infectious diseases that humanity continues to experience.[29] The occurrence of multi-drug resistance and extremely-drug resistant strains of Mycobacterium tuberculosis made the situation worse; therefore, an urgent solution to combat TB is required. Phage therapy is getting more attention from researchers around the world and can be a promising alternative to tackle the rising drug resistance problem in M. tuberculosis. Their natural capacity to kill Mycobacterium spp. has attracted researchers to work on phages for the treatment of tuberculosis.[30-32] Interestingly, in the 1970s such phages were utilized for typing of clinical strains of M. tuberculosis in different epidemiological studies in Europe and Asia.[33] Two Mycobacterium phages (VIT001 and VIT002) were isolated from the soil samples collected from Government Vellore Medical College & Hospital using M. smegmatis mc2155 as the host. Both the phages exhibited clear plaques with a size of approximately 1.2 ± 0.04 mm and 3.4 ± 0.27 mm diameter respectively, in double agar overlay method. TEM analysis showed that both the phages belong to Siphoviridae family (Fig.6). Mycobacterium phage VIT001 showed an interesting morphology containing prolate head of 83 ± 1.07 nm in length and 32.4 ± 0.18 nm in width with a long tail of about 201 ± 2.28 nm in length. In contrast to this, Mycobacterium phage VIT002 showed an icosahedral head of size 54 ± 0.1 nm and a long non-contractile tail of about 225.47 ± 1.67 nm (Fig.6).
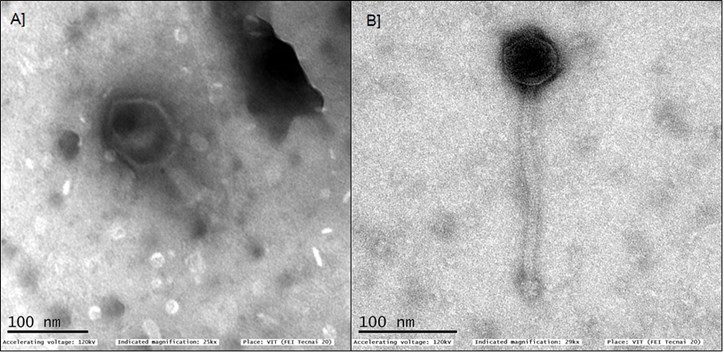
Figure 8: TEM images of bacteriophages stained using 2% [w/v] phosphotungstic acid (A) and 2% [w/v] uranyl ace- tate (B). A) Escherichia phage- Myoviridae; B) Mycobacteriophage- Siphoviridae
Mycobacteriophages which infects the Mycobacterium spp. were grouped into 29 (A-Z) clusters and ten singletons in the actinobacteriophage database (phagesdb.org) and each cluster has been further subdivided into sub-clusters based on the sequence nucleotide identity and gene content. The database shows cluster I and O to have mycobacteriophages with prolate heads. India has amongst the highest occurrence of tuberculosis infection rate in the world. Therefore, there is a possibility for the widespread presence of the bacterial host i.e. Mycobacterium in the environment indicating the presence of varied mycobacteriophages. However, there are minimal reports about the mycobacteriophages in India.[34] We believe that finding novel mycobacteriophages would not only contribute in expanding the existing database, it may also lead to the identification of unexplored virulent phages, with promising therapeutic potential.
Bacteriophages against pathogenic E. coli were isolated from Ganges river water collected from Varanasi and municipal sewage water collected from Karur. More than 10 Escherichia phages were isolated from Ganges river water against 16 different E. coli strains. Only one Escherichia phage was further characterized for therapy mainly based on their broad-host-range infectivity.[13] The complete characterization of Escherichia phage myPSH1131 isolated from Ganges river water was studied in detail and has been published previously.[13] This phage was found to form clear plaques in the double agar overlay plates. Historically, in the early 1890s, bacteriophages were first isolated from Ganges river water and reports describe the Ganges river water’s “antibacterial properties” and the presence of bacteriophages against bacterial pathogens.[3,37] Another Escherichia phage myPSH2311 was isolated from sewage water samples; judging from its morphology, this phage belongs to the Siphoviridae family with an icosahedral head of 33 ± 3.0 nm and a non-contractile tail of 65 ± 2.5 nm in length. An extensive characterization of the phage and therapeutic potential was published previously.[12] From the sewage water samples, more than 25 bacteriophages were isolated against pathogenic E. coli in this study.
Klebsiella pneumoniae is an opportunistic pathogen that can cause severe infections in hospitalized patients. Recent reports on infections caused by carbapenem-resistant K. pneumoniae are alarming, and alternative clinical strategies such as phage therapy might present a treatment option.[10,38] The isolation of virulent phages with broad-host-range activity is challenging and the source of the sample plays an essential role in effective phage isolation. In this study, Klebsiella phage with broad-host-range infectivity was isolated from sewage water samples collected from sewage water samples collected in Karur. The Klebsiella phage was found to form clear plaques surrounded by a halo and morphologically belongs to the Podoviridae family with an icosahedral head of 80 ± 4.5 nm and a very short non-contractile tail. A comprehensive characterization of Klebsiella phage myPSH1235 has been published previously.[12]
The isolated Enterobacter phage myPSH1140 was found to belong to the Myoviridae family with the icosahedral head of 80 ± 2.0 nm and long contractile tail with tail fibers of about 101 ± 3.5 nm in length. The characteristics of the phage have been described elsewhere.[12] The phage has been isolated from municipal sewage water in the Indian city of Chennai. Though the sewage water samples are known to carry many bacteriophages (against different bacterial strains), the selection of specific phage is essential for therapeutic applications.[12,13] Under certain conditions, the phage cocktails are used in sewage treatment plants to remove specific groups of bacteria; even in this instance, the use of virulent phages should be preferred to eliminate the transmission of resistance genes by lysogenic phages.[39]
In this study, most of the phages were isolated from sewage water samples. Earlier reports also described the abundance of bacteriophages in sewage water samples.[40,41] The most abundant phages in the environment are known to belong to Siphoviridae family and we report nine Siphoviruses, two Podoviruses and one Myovirus. Many Siphoviruses exhibit a lysogenic cycle such as phage lambda.[42] However, as the use of virulent phages is considered most suitable for therapy, it is important to characterize the phages. Therefore, based on several microbiological tests, including plaque morphology (on double agar overlay plate), virulent phages can be identified. A halo-clear plaque formed by the virulent phages can be differentiated from the turbid, bulls-eyed plaques that are often formed by temperate phages (Fig.7).
“Halo-clear plaques” are characterized by the occurrence of a defined boundary with a clear-cut edge, while “bulls-eye plaques” are turbid and have fizzy edges. The plaques formed by virulent / lytic phages look clear due to the complete lysis of the bacterial cells; whereas temperate / lysogenic phages form rings of bacterial growth around the clear regions (lysis) due to only partial killing of the host (Fig.7). This simple virulent phage identification method can be used to screen phages for clinically important pathogens and during phage screening under time constraint. Except for few phages such as T7, most phage plaques presume certain size and form a boundary, either clear-cut or fizzy edges.[43] The plaque morphology was also found to be differed based on virion structure or morphology.[43] Previously, we reported that plaque morphologies of different phages depended on the microbiological medium used.[21]
For TEM imaging, a simpler phage filtration technique was found to be most effective compared to precipitation and purification. Both precipitation and purification takes longer and is a tedious process, therefore, a simple phage filtration using 0.22 micron syringe filters are sufficient for TEM imaging. We also observed that samples containing precipitated phages often showed unwanted crystal structure formation, obscuring the viruses during imaging which might be due to the salts present in the solutions. In order to avoid problems during imaging, we recommend using filtered phages for the TEM. Regarding staining of the particles, the use of 2% phosphotungstic acid the TEM images were dark even though the plaque morphology was observed around the black background (Fig.8A). In comparison, 1% [w/v] uranyl acetate served as a good staining agent (Fig.8B). Though there are numerous studies that are focused on phage biology and phage characterization for therapy, there is no simple and effective technique for virulent phage identification. Even though phage life cycle and genomic analysis are considered to be important to characterize therapeutic phages, this simple identification technique can be used to screen virulent phages for analysis.
Bacteriophage research is gaining more and more attention due to vast applications, in particular phage therapy to cure bacterial infections in the antibiotic-resistant era. This study details a simple method for the isolation and identification of virulent phages by using plaque morphology and TEM analysis. Also, an abundance of bacterio- phages in the sewage system was observed, which is an excellent source for phage isolation. This study contributes to the effort to describe simple techniques to facilitate the rapid characterization of virulent phages for potential therapeutic use.
The authors would like to thank the Vellore Institute of Technology for providing 'VIT SEED grant'. We would also like to thank VIT TEM facility for providing Transmission Electron Microscopic images.
The authors declare that there is no conflict of interest.
1. Keen EC. A century of phage research: bacteriophages and the shaping of modern biology. Bioessays. 2015; 37(1):6-9.
2. Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brüssow H. Phage-host interaction: an ecological perspective. J bacterial. 2004; 186(12):3677-86.
3. Manohar P, Tamhankar AJ, Leptihn S, Ramesh N. Pharmacological and Immunological Aspects of Phage Therapy. Infec- tious Microbes & Diseases. 2019; (2):34-42.
4. Loh B, Leptihn S. A Call For a Multidisciplinary Future of Phage Therapy to Combat Multi-drug Resistant Bacterial Infections. Infectious Microbes & Diseases. 2020; 2(1):1-2
5. Aslam S, Schooley RT. What’s Old Is New Again: Bacteriophage Therapy in the 21st Century. Antimicrob Agents Chemother. 2019; 64(1):e01987.
6. Luong T, Salabarria AC, Roach DR. Phage therapy in the resistance era: where do we stand and where are we going?. Clin Ther. 2020; 42(9):1659-1680.
7. Fortier LC, Sekulovic O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence. 2013; 4 (5): 354-65.
8. Loh B, Chen J, Manohar P, Yu Y, Hua X and Leptihn S. A Biological Inventory of Prophages in A. baumannii Genomes Reveal Distinct Distributions in Classes, Length, and Genomic Positions. Front Microbiol. 2020; 11:579802.
10. Manohar P, Loh B, Athira S, Nachimuthu R, Hua X, Welburn SC, Leptihn S. Secondary Bacterial Infections During Pulmonary Viral Disease: Phage Therapeutics as Alternatives to Antibiotics?. Front Microbiol. 2020; 11:1434.
11. Adriaenssens EM, Sullivan MB, Knezevic P, van Zyl LJ, Sarkar BL, Dutilh BE, Alfenas-Zerbini P, Łobocka M, Tong Y, Brister JR, Switt AI. Taxonomy of prokaryotic viruses: 2018-2019 update from the ICTV Bacterial and Archaeal Viruses Subcommittee. Arch Virol. 2020; 165:1253-1260.
12. Manohar P, Tamhankar AJ, Lundborg CS, Nachimuthu R. Therapeutic characterization and efficacy of bacteriophage cocktails infecting Escherichia coli, Klebsiella pneumoniae, and Enterobacter species. Front Microbiol, 2019; 10:574.
13. Manohar P, Tamhankar AJ, Lundborg CS, Ramesh N. Isolation, characterization and in vivo efficacy of Escherichia phage myPSH1131. PloS one. 2018; (10):e0206278.
14. Royam MM, Nachimuthu R. Isolation, characterization, and efficacy of bacteriophages isolated against Citrobacter spp. an in vivo approach in a zebrafish model (Danio rerio). Res Microbiol. 2020; S0923-2508 (20): 30082-6.
15. Manohar P, Nachimuthu R, Lopes BS. The therapeutic potential of bacteriophages targeting gram-negative bacteria using Galleria mellonella infection model. BMC Microbiol. 2018; 18(1):97.
16. Ramesh N, Archana L, Royam MM, Manohar P, Eniyan K. Effect of various bacteriological media on the plaque morphology of Staphylococcus and Vibrio phages. Access Microbiol. 2019; 1(4):e000036.
17. Manohar P, Loh B, Leptihn S. Will the Overuse of An- tibiotics During the Coronavirus Pandemic Accelerate Antimicrobial Resistance of Bacteria? Infectious Microbes & Diseases. 2020; 2(3): 87-88.
18. Manohar P, Ramesh N. Improved lyophilization conditions for long-term storage of bacteriophages. Sci Rep. 2019; 9(1):1-0.
19. Loh B, Gondil VS, Manohar P, Khan FM, Yang H, Leptihn S. Encapsulation and Delivery of Therapeutic Phages. Appl Environ Microbiol. 2020
20. Nobrega FL, Vlot M, de Jonge PA, Dreesens LL, Beau- mont HJ, Lavigne R, Dutilh BE, Brouns SJ. Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol. 2018; 16(12): 760-73.
21. Ramesh N, Archana L, Royam MM, Manohar P, Eniyan K. Effect of various bacteriological media on the plaque morphology of Staphylococcus and Vibrio phages. Access Microbiol. 2019; 1(4):e000036.
22. Merkier AK, Rodríguez MC, Togneri A, Brengi S, Osuna C, Pichel M, Cassini MH, Centrón D. Outbreak of a cluster with epidemic behavior due to Serratia marcescens after colistin administration in a hospital setting. J Clin Microbiol. 2013; 51(7):2295-302.
23. Fleisch F, Zimmermann-Baer U, Zbinden R, Bischoff G, Arlettaz R, Waldvogel K, Nadal D, Christian R. Three consecutive outbreaks of Serratia marcescens in a neonatal intensive care unit. Clin Infect Dis. 2002; 34 (6):767-73.
24. Duman M, Saticioglu IB, Buyukekiz AG, Balta F, Altun S. Molecular characterization and antimicrobial resistance profile of atypical Citrobacter gillenii and Citrobacter sp. isolated from diseased rainbow trout (Oncorhynchus mykiss). J Glob Antimicrob Res. 2017; 10:136e42.
25. Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, Holland TL, Fowler VG. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol. 2019; 17(4):203-18.
26. Feng T, Leptihn S, Dong K, Loh B, Zhang Y, Li M, Guo X, Cui Z. Characterisation of bacteriophage JD419, a Staphylococcal phage with an unusual morphology and broad host range. bioRxiv. PREPRINT. 2020.
27. Capparelli R, Parlato M, Borriello G, Salvatore P, Iannelli D. Experimental phage therapy against Staphylococcus aureus in mice. Antimicrob Agents Chemother. 2007; 51(8):2765-73.
28. Kifelew LG, Warner MS, Morales S, Thomas N, Gordon DL, Mitchell JG, Speck PG. Efficacy of Lytic Phage Cocktails on Staphylococcus aureus and Pseudomonas aeruginosa in Mixed-Species Planktonic Cul- tures and Biofilms. Viruses. 2020; 12(5):559.
29. Knight GM, McQuaid CF, Dodd PJ, Houben RM. Global burden of latent multidrug-resistant tuberculosis: trends and estimates based on mathematical model- ling. The Lancet Infect Dis. 2019; 19(8): 903-12.
30. Chatterjee S, Mitra M, Das Gupta SK. A high yielding mutant of mycobacteriophage L1 and its application as a diagnostic tool. FEMS microbiol letters. 2000;188 (1):47-53.
31. Broxmeyer L, Sosnowska D, Miltner E, Chacón O, Wagner D, McGarvey J, Barletta RG, Bermudez LE. Killing of Mycobacterium avium and Mycobacterium tuberculosis by a mycobacteriophage delivered by a nonvirulent mycobacterium: a model for phage therapy of intracellular bacterial pathogens. J Infect Dis. 2002; 186(8):1155-60.
32. Danelishvili L, Young LS, Bermudez LE. In vivo efficacy of phage therapy for Mycobacterium avium infec- tion as delivered by a nonvirulent mycobacterium. Microb Drug Resist. 2006;12(1):1-6.
33. McNerney R. TB: the return of the phage. A review of fifty years of mycobacteriophage research. The Interna- tional Journal of Tuberculosis and Lung Disease. 1999; 3(3):179-84.
34. Pope WH, Jacobs-Sera D, Russell DA, Peebles CL, Al- Atrache Z, Alcoser TA, Alexander LM, Alfano MB, Alford ST, Amy NE, Anderson MD. Expanding the diversity of mycobacteriophages: insights into genome architecture and evolution. PloS one. 2011; 6 (1):e16329.
35. Pope WH, Bowman CA, Russell DA, Jacobs-Sera D, Asai DJ, Cresawn SG, Jacobs Jr WR, Hendrix RW, Lawrence JG, Hatfull GF. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. elife. 2015; 4:e06416.
36. Bajpai U, Mehta AK, Eniyan K, Sinha A, Ray A, Virdi S, Ahmad S, Shah A, Arora D, Marwaha D, Chauhan G. Isolation and characterization of bacteriophages from India, with lytic activity against Mycobacterium tuberculosis. Can J Microbiol. 2018; 64(7):483-91.
37. Khairnar K. Ganges: special at its origin. Journal of Biological Research-Thessaloniki. 2016; 23(1):1-2.
38. Asavarut P, Hajitou A. The phage revolution against antibiotic resistance. The Lancet Infect Dis. 2014; 14 (8):686.
39. Wu B, Wang R, Fane AG. The roles of bacteriophages in membrane-based water and wastewater treatment processes: A review. Water res. 2017; 110:120-32.
40. Jurczak-Kurek A, Gąsior T, Nejman-Faleńczyk B, Bloch S, Dydecka A, Topka G, Necel A, Jakubowska- Deredas M, Narajczyk M, Richert M, Mieszkowska A. Biodiversity of bacteriophages: morphological and biological properties of a large group of phages isolated from urban sewage. Sci Rep. 2016;6(1):1-7.
41. Muniesa M, Imamovic L, Jofre J. Bacteriophages and genetic mobilization in sewage and faecally polluted environments. Microb Biotechnol. 2011;4(6):725-34.
42. Hyman P, Abedon S.T. Bacteriophage (overview), Edi- tor(s): Schaechter M. Encyclopedia of Microbiology (Third Edition), Academic Press, 2009, 322-338
Prasanth M, Kandasamy E, Loganathan A, Madurantakam R. M, Sebastian L, Nachimuthu R. Morphological characterization techniques for the isolation of virulent bacteriophages from environmental sources. Int J Bacteriophage Res 2021:1:1-9.